class: center, middle, inverse, title-slide .title[ # Lab1: Introduction to the R language, R environment, RStudio, and R Markdown ] .subtitle[ ## Data types, data structures, subsetting, functions, workflow, markdown, and R Markdown ] .author[ ### Christopher Moore ] --- # Introduction to R(1 + Studio + Markdown) 4. **Workflow**: locating and devising a systematic way of working 5. **Markdown**: modifying plain text 6. **R Markdown Document**: a "dynamic document" that allows you to streamline your workflow 3. **Functions**: the heaRt of it all (❤️) 0. **Data types**: categories of data that R uses and interprets 1. **Data structures**: how R organizes different types of data; i.e., perhaps the most important thing about R that you never thought too deeply about 2. **Subsetting**: *very* useful for data analysis. For this course, we're creating all of our data *de novo*, so we won't necessarily be subsetting as much as other courses. --- class: inverse, center, middle # workflows ---  # we all need to work on our workflows --- # Workflows: directories When logging on a computer, the home directory is: *"hard drive"/"users"/"logged in user"* ```r /Users/`userID` # generally /Users/cmmoore # on *my* computer ``` Here you see `Applications`, `Desktop`, `Documents`, `Downloads`, etc. -- *** When logging on to RStudio server, the home directory is: *"colby's hard drive/users/courses/bi382/logged in user"* ``` /Volumes/Courses/BI382/`userID` # generally /Volumes/Courses/BI382/cmmoore ``` Here you see one directory: `Private` --- # Workflow: directories Create the following directories in `Private`: * `LabNotebook` * `ProblemSets` * `RemodelingProject` I would: 1. Make a new folder in `/Volumes/Courses/BI382/"userID"` called `Work` (or something) 2. Copy this structure into `/Volumes/Courses/BI382/"userID"/Work` --- # Check working directory `getwd()` with no argument --- class: inverse, center, middle # R Markdown --- class: inverse, center, middle # Data types in R --- # Data types in R 1. character 2. numeric 3. integer 4. logical 5. complex --- # Data types in R ## 1. character: `"a"` or `"I like turtles"` ```r is.character("Chris") ## [1] TRUE paste0("file", ".ext") ## [1] "file.ext" str("function") ## chr "function" ``` --- # Data types in R ## 2. numeric: `1` or `-4` or `16^(1/2)` ```r is.numeric(42) ## [1] TRUE 2 + 4 / 5 # order of operations is valid ## [1] 2.8 4*3^2 # order of operations is valid ## [1] 36 str(4) ## num 4 ``` --- # Data types in R ## 3. integer: `3L` ```r is.integer(3L) ## [1] TRUE 1L + 2L ## [1] 3 str(11L) ## int 11 ``` --- # Data types in R ## 4. logical: `T` or `F` (equivalent to `TRUE` and `FALSE`, respectively) ```r is.logical(T) ## [1] TRUE is.logical(D) ## [1] FALSE 1 == 1 ## [1] TRUE 1 == 2 ## [1] FALSE str(2 == 2) ## logi TRUE ``` --- # Data types in R ## 5. complex: ```r is.complex(4) ## [1] FALSE is.complex(4i) ## [1] TRUE complex(real = 3, imaginary = 2) + complex(real = 1, imaginary = 20) ## [1] 4+22i str(3 + 2i) ## cplx 3+2i ``` --- class: inverse, center, middle # Data structures in R --- # Data structures in R Data structures differ by the number of dimensions and whether the data types are the same or different d | Homogeneous | Heterogeneous :----:|:------------------:|:------------: 1d | atomic vector | list `\(^\dagger\)` 2d | matrix | dataframe *n*d | array | --- # Data structures in R: 1d * homogenous input, homogeneous output ```r c(1, 2, 3) ## [1] 1 2 3 str(c(1, 2, 3)) ## num [1:3] 1 2 3 ``` * heterogeneous input, homogeneous output ```r c(F, 18L, 4) ## [1] 0 18 4 str(c(F, 18L, 4)) ## num [1:3] 0 18 4 ``` --- # Data structures in R: 1d * heterogeneous input, heterogeneous output ```r list(F, 18L, 4) ## [[1]] ## [1] FALSE ## ## [[2]] ## [1] 18 ## ## [[3]] ## [1] 4 str(list(F, 18L, 4)) ## List of 3 ## $ : logi FALSE ## $ : int 18 ## $ : num 4 ``` --- # Data structures in R: 2d * matrix: a homogeneous data structure ```r matrix(data = c(1, 2, 3, 4), nrow = 2, ncol = 2) ## [,1] [,2] ## [1,] 1 3 ## [2,] 2 4 matrix(data = c(1, 2, FALSE, 4), nrow = 2, ncol = 2) ## [,1] [,2] ## [1,] 1 0 ## [2,] 2 4 ``` --- # Data structures in R: 2d * dataframe: a concatenation of atomic vectors as the columns ```r data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F) ## c.1..2..3. c..a....b....c.. c..d....e...F. ## 1 1 a d ## 2 2 b e ## 3 3 c FALSE str(data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F)) ## 'data.frame': 3 obs. of 3 variables: ## $ c.1..2..3. : num 1 2 3 ## $ c..a....b....c..: chr "a" "b" "c" ## $ c..d....e...F. : chr "d" "e" "FALSE" ``` * we are punished when we have atomic vectors that are different lengths ```r data.frame(c(1, 2, 3), c("a", "b", "c"), c(T, T, T, F)) ## Error in data.frame(c(1, 2, 3), c("a", "b", "c"), c(T, T, T, F)): arguments imply differing number of rows: 3, 4 ``` --- # Data structures in R: *n*d * array: a homogeneous data structure ```r array(data = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10:18), dim = c(2, 3, 3)) ## , , 1 ## ## [,1] [,2] [,3] ## [1,] 1 3 5 ## [2,] 2 4 6 ## ## , , 2 ## ## [,1] [,2] [,3] ## [1,] 7 9 11 ## [2,] 8 10 12 ## ## , , 3 ## ## [,1] [,2] [,3] ## [1,] 13 15 17 ## [2,] 14 16 18 ``` --- # Data structures in R: a note on lists `\(^\dagger\)` * lists are 1d, but each element can take any structure in R ```r list("a", c("a", "b"), matrix(data = c(1, 2, 3, 4), nrow = 2, ncol = 2)) ## [[1]] ## [1] "a" ## ## [[2]] ## [1] "a" "b" ## ## [[3]] ## [,1] [,2] ## [1,] 1 3 ## [2,] 2 4 ``` --- # A visual metaphor of lists <blockquote>Indexing lists in R. "Inspired by the Residence Inn" — Hadley Wickham (@hadleywickham) <a href="https://twitter.com/hadleywickham/status/643381054758363136?ref_src=twsrc%5Etfw">September 14, 2015</a></blockquote> <script async src="https://platform.twitter.com/widgets.js" charset="utf-8"></script> 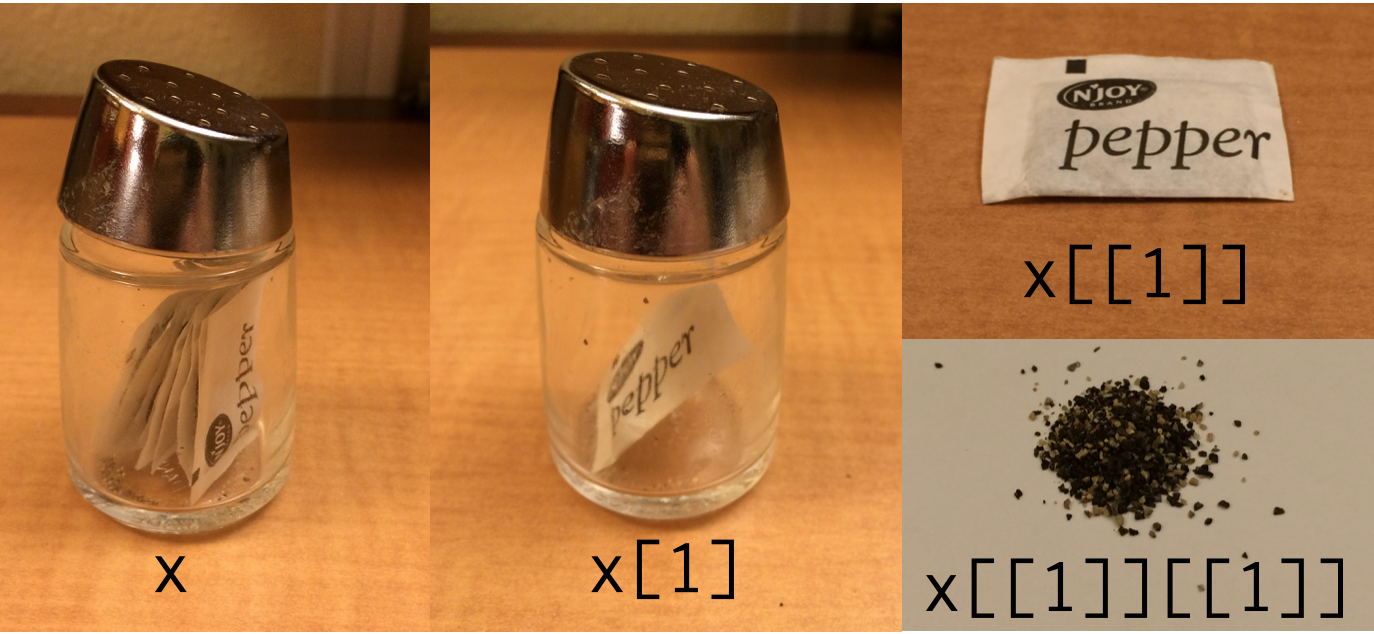 --- class: inverse, center, middle # Subsetting --- # Subsetting: dimensionality Accessing your data structure depends on its dimensionality * 1d structures: 1 or more numbers or conditions that correspond to the position of the element(s) in which you're interested * All structures can take logical values---YAY!!! ```r c(10, 20, 30, 40)[3] ## [1] 30 c(10, 20, 30, 40)[c(2, 4)] ## [1] 20 40 c(10, 20, 30, 40)[c(F, T, T, F)] ## [1] 20 30 list(1, F, 3)[3] ## [[1]] ## [1] 3 ``` --- # Subsetting: dimensionality Accessing your data structure depends on its dimensionality * 2d structures: 1 or more numbers or conditions that correspond to the position of the element(s) in which you're interested * Remember it's always **[rows, columns]** * matrices ```r matrix(data = c(10, 20, 30, 40), nrow = 2, ncol = 2) ## [,1] [,2] ## [1,] 10 30 ## [2,] 20 40 matrix(data = c(10, 20, 30, 40), nrow = 2, ncol = 2)[1,] ## [1] 10 30 matrix(data = c(10, 20, 30, 40), nrow = 2, ncol = 2)[,1] ## [1] 10 20 matrix(data = c(10, 20, 30, 40), nrow = 2, ncol = 2)[1,2] ## [1] 30 ``` --- # Subsetting: dimensionality Accessing your data structure depends on its dimensionality * 2d structures: 1 or more numbers or conditions that correspond to the position of the element(s) in which you're interested * Remember it's always **[rows, columns]** * dataframes ```r data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F) ## c.1..2..3. c..a....b....c.. c..d....e...F. ## 1 1 a d ## 2 2 b e ## 3 3 c FALSE data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F)[2, 2] ## [1] "b" data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F)$c.1..2..3. ## [1] 1 2 3 data.frame(c(1, 2, 3), c("a", "b", "c"), c("d", "e", F), stringsAsFactors = F)$c.1..2..3.[2] ## [1] 2 ``` --- # Subsetting: logical values Logical values will allow you to use conditional statements you to subset data ```r 1 == 1 ## [1] TRUE 1 < 2 ## [1] TRUE c(10, 20, 30, 40)[c(T, F, T, F)] ## [1] 10 30 c(10, 20, 30, 40) > 10 ## [1] FALSE TRUE TRUE TRUE c(10, 20, 30, 40)[c(10, 20, 30, 40) > 10] ## [1] 20 30 40 c(10, 20, 30, 40)[c(10, 20, 30, 40) == 30] ## [1] 30 c(10, 20, 30, 40)[c(10, 20, 30, 40) %% 4 == 0] # %% is the modulus, which is the remainder in division ## [1] 20 40 ``` --- # Subsetting: logical values Logical operators |Operator | Description | |:--------------:|:-------------------------:| | `<` | less than | | `<=` | less than or equal to | | `>` | greater than | | `>=` | greater than or equal to | | `==` | is exactly equal to | | `!=` | is not equal to | | `!x` | is not `x` | | `x `|` y` | `x` or `y` | | `x&y` | `x` and `y` | --- # Subsetting: IDing the elements you want to subset `which()` identifies which elements of a logical data structure are `T` (you can also choose to ID `F` instead) ```r c(10, 20, 30, 40) > 10 # conditional statement ## [1] FALSE TRUE TRUE TRUE which(c(10, 20, 30, 40) > 10) # using `which()` ## [1] 2 3 4 c(10, 20, 30, 40)[c(10, 20, 30, 40) > 10] # subsetting ## [1] 20 30 40 c(10, 20, 30, 40)[which(c(10, 20, 30, 40) > 10)] # subsetting ## [1] 20 30 40 ``` --- class: inverse, center, middle # functions --- # Functions As intrdocuted, the heaRt of R. *Every*thing that has parentheses—e.g., `c()`, `which()`, `matrix()`—is a function. -- <p align="center"> <img src="https://upload.wikimedia.org/wikipedia/commons/thumb/3/3b/Function_machine2.svg/1200px-Function_machine2.svg.png" alt="Drawing" style="width: 300px;"/> </p> --- # Functions Three parts of functions in R 1. **formals**: a list of the *arguments* (the inputs of the function) 2. **body**: the code inside of the function 3. **environment**: beyond the scope of this course, but "maps" functions' variables --- # Functions Writing a function ```r (function(x) x^2) # this is a function ## function(x) x^2 ``` -- Running a function ```r (function(x) x^2)(x = 3) # add parentheses and it evaluares the argument `x = 3` ## [1] 9 ``` -- Let's save the function to memory using the assignment operator `<-`, then run it ```r my.fun <- function(x) x^2 my.fun(x = 2) ## [1] 4 ``` --- # Functions Pseudocode ``` function name <- function(arguments) { code to be executed return(list of variables to be returned) } ``` -- A real function ```r cuboid.vol <- function(l, w, h) { plane.area <- l*w total.vol <- plane.area*h return(total.vol) } ``` -- Run it ```r cuboid.vol(l = 2, w = 5, 9) # arguments are evalued in order if not specified ## [1] 90 ```